We study macromolecular complexes with enzymatic function involved in RNA metabolism and in the regulation of gene expression. RNA plays myriad roles in eukaryotic cells, with its adaptability partly due to its low-diversity chemical composition, which allows the polymer to change structure depending on the environment, binding partners and co-factors. Our current projects include RNP (ribonucleoprotein) enzymes that introduce chemical modifications on various types of RNA, as well as RNP complexes involved in the biogenesis of piRNA, a class of regulatory RNA responsible for transposon regulation in germ-line cells.
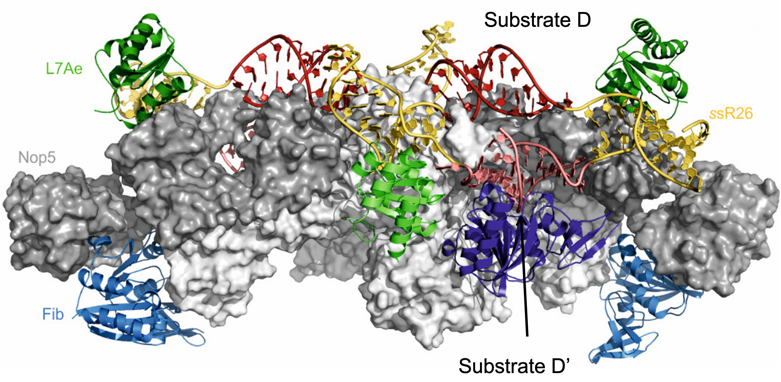
In the past, we have applied the integrative structural biology approach to solve the structure of the 2’-O-methylation enzyme, a complex of 12 proteins and 10 RNAs, which methylates the 2’-O position of ribosomal RNA (rRNA) in the nucleolus. We not only determined its three-dimensional architecture in solution (Lapinaite et al., Nature, 2013; see figure), but also discovered how dynamic processes, which are dependent on the primary sequence of the substrate RNA, regulate the efficiency of methylation at different rRNA sites (Graziadei et al., eLife, 2020).
Currently, we aim to develop the RNA modification machinery as a therapeutic target in the treatment of disease. Defects in RNA modification patterns have been shown to cause disease in humans; furthermore, some pathogens are require extensive editing of their mRNA to yield functional protein expression machinery. The biological roles of many RNA modifications are not yet fully understood, with proposed mechanisms ranging from structure (de)stabilization, protection from hydrolysis, and protein binding. Through our structural studies, we seek to understand the mechanisms by which RNA modifications are achieved and regulated, with the aim of establishing RNA-editing enzymes as drug targets.
In the piRNA field, we have used NMR spectroscopy to study the dynamics of the DEAD-box helicase protein Vasa through the unwinding reaction, namely in complexes with ADP/ATP, single-stranded RNA (ssRNA)/double-stranded RNA (dsRNA) and combinations thereof. DEAD-box helicases use ATP to unwind short double-stranded RNA. Following ATP binding to the N-terminal domain of the two-domain helicase core, these two core domains are thought to close upon each other, forming the dsRNA binding surface and stimulating ATP hydrolysis. However, because the crystallographic structures of DEAD-box helicases in complex with either ADP- or ATP-analogues are identical and the complex of a DEAD-box helicase with ATP and dsRNA could not yet be isolated, it has remained unclear how ATP (but not ADP) promotes dsRNA unwinding. We have used NMR chemical shift perturbations (CSPs) and relaxation-dispersion profiles of the DEAD-box helicase Vasa in the presence of ADP or ATP, and ssRNA or dsRNA, to demonstrate that the conformations of the ATP- and ADP-bound forms of the N-terminal domain are different, and have proposed a mechanism by which subtle changes in the tertiary structure facilitate cycling of Vasa through the unwinding reaction. In addition, we have identified the existence of distinct, co-existing protein–RNA–nucleotide complexes that may support an alternative cellular function of Vasa, in which the protein acts either as an RNA clamp rather than a helicase.
We are also studying the role and the structure of the protein Qin, a multidomain protein consisting of an N-terminal RING domain and five Tudor-like domains, which is involved in the Ping-Pong cycle during secondary piRNA biogenesis.
