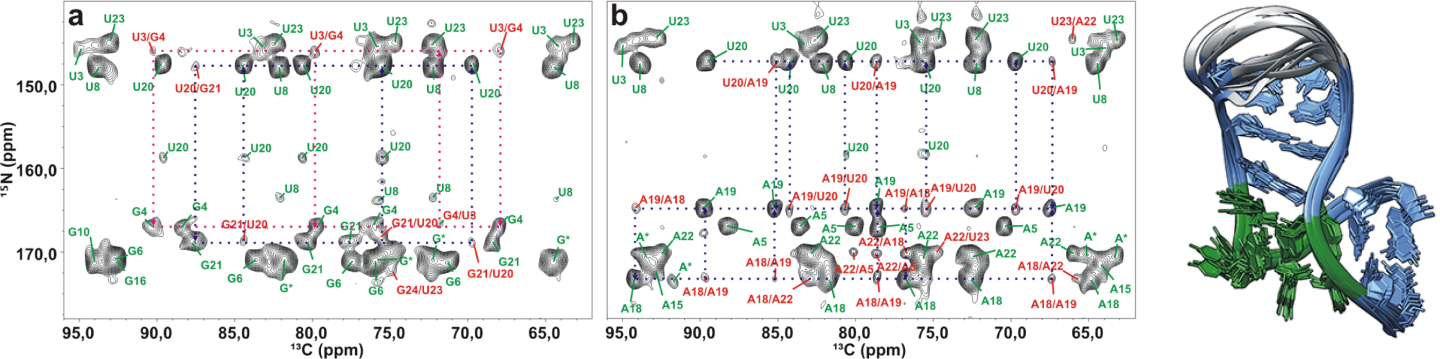
We have pioneered the application of solid-state NMR (ssNMR) to RNA and RNPs. ssNMR is particularly suitable for RNA as part of large complexes, because the quality of ssNMR spectra is independent of the size of the molecule. In addition, ssNMR does not require crystals, and the sample preparation process preserves a high level of hydration, which is essential to support proper native folding of RNA.
We have determined the first de novo high-resolution structure of RNA by ssNMR (Marchanka et al., Angewandte Chemie, 2013; Marchanka et al., Nature Communications, 2015; Figure 1) and have developed methods for proton-detected ssNMR of RNA and identification of base-pairing patterns (Marchanka et al., Chemical Communications, 2018). Furthermore, we have determined the first NMR structure of an RNA–protein complex using only ssNMR-derived data. We believe that there remains significant unexplored potential for ssNMR of RNA and RNPs, which we are exploiting for both methodological developments and novel areas of application.