Molecules establish networks of physical interactions to perform a plethora of highly regulated tasks, such as energy storage and usage, waste disposal and recycling, protection against external attacks, and many others. To establish these networks, molecules bind to each other through their complementary shapes, in a process figuratively described as "lock-and-key". In recent years, this concept has been extended to include the notion that some molecules do not have a well-defined shape but are rather malleable; the shape of these molecule can adapt to establish interactions with other molecules of more rigid, pre-defined three-dimensional structures. This mode of interaction massively increases the scope of intermolecular communication, and provides opportunities for internal and external regulation by influencing the malleability of the adaptable molecules. Even more recently, efficient and functional interactions have been demonstrated to exist between spaghetti-like molecules, which are able to bind tightly to each other while preserving their disordered nature.
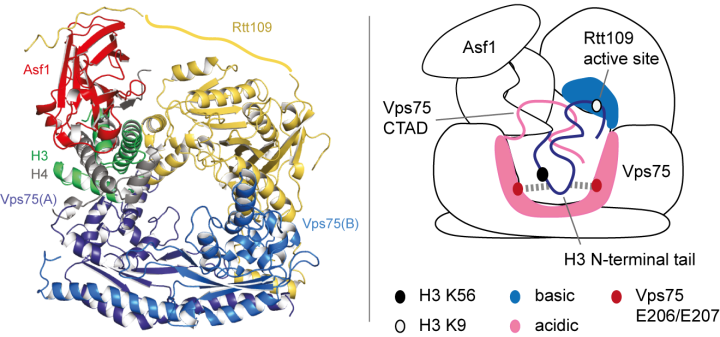
The paradigm of “(folded) structure-function relationships” has been challenged by a revolutionary paper (Borgia et al., Nature, 2018) that describes a functional complex between two intrinsically unfolded proteins, which both remain disordered upon interaction (the two-rope-tangle model). In some of our recent work, we have found a second example of this type of interaction (Danilenko et al., Nature Communications, 2019). Here, to understand how histone chaperones Asf1 and Vps75 together promote H3 K9-acetylation, we derived a solution-state structural model of the acetyltransferase enzyme Rtt109 in complex with Asf1 and Vps75 and the histone dimer H3:H4 using a combination of NMR spectroscopy, small-angle neutron scattering (SANS) and molecular modelling (Figure 1, left). We showed that Vps75 promotes K9-acetylation by engaging the H3 N-terminal tail in fuzzy electrostatic interactions with its disordered C-terminal domain, thereby confining the H3 tail to a wide central cavity faced by the Rtt109 active site (Figure 1, right). These fuzzy interactions between disordered domains achieve localization of lysine residues in the H3 tail to the catalytic site with minimal loss of entropy and may represent a common mechanism for enzymatic reactions involving highly disordered substrates.
We believe that this apparently unusual interaction mode is much more common than anticipated and may represent a general mechanism for chaperoning disordered substrates. We are using integrative structural biology and advanced molecular modelling to study at the atomic-level how disordered regions of chaperones interact with disordered substrates and present them to the catalytic site of enzymes. This work goes beyond the conventional structure determination process, as the concept of a well-defined complex structure is substituted by a large ensemble of transient conformations, a scenario that may promote a favourable balance between the enthalpic and entropic terms of the free-energy of binding. Given the abundance of disordered motifs in the eukaryotic proteome, this research promises to significantly improve our understanding of many eukaryotic cellular processes.
